Erika McCarthy1, Şölen Ekesan1, and Darrin M. York1
1Laboratory for Biomolecular Simulation Research, Institute for Quantitative Biomedicine and Department of Chemistry and Chemical Biology, Rutgers University, Piscataway, NJ 08854, USA
In this tutorial you will utilize antechamber to determine atom types carry out the restrained electrostatic potential (RESP) procedure to obtain partial charges for the non-standard residue GUN, O6 methylated guanine ligand), and build parameter files. You will go through the following sections:
The necessary inputs and subsequent outputs for all parts of this tutorials are available in:
/expanse/projects/qstore/amber_ws/tutorials/Param_nonstandard_residue_MTR1/input
and
/expanse/projects/qstore/amber_ws/tutorials/Param_nonstandard_residue_MTR1/output
The inputs should be copied to your working directory. If you unable to produce an output for a given step, copy it from the output directory in order to proceed.
To use AMBER programs, you must:
module use /expanse/projects/qstore/amber_ws/packages/modulefiles/
module load workshop/software.
[user@cluster] mkdir Param_nonstandard_residue_MTR1
[user@cluster] cd Param_nonstandard_residue_MTR1
[user@cluster] cp /expanse/projects/qstore/amber_ws/tutorials/Param_nonstandard_residue_MTR1/input ./AMBER residue parameters are defined by atom types and partial charges. AMBER has a wide list of atom types with established MM parameters regarding bond lengths, angles, dihedral angles and vdW radii. Partial charges that go into the electrostatics calculation of the force are described separately. So generating parameters for a new residue requires atom type selection and partial charge determination.
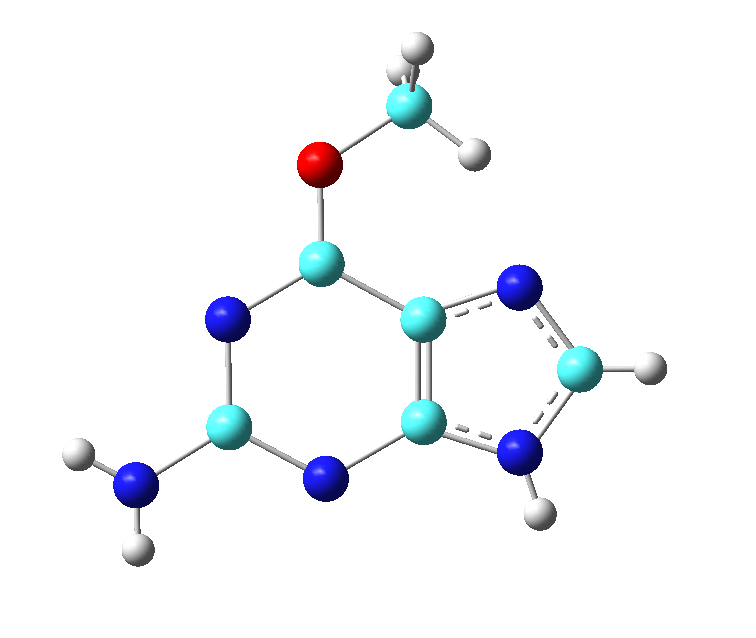
In this section you will see how a Gaussian input file was created using GaussView based on the ligand structure in the pdb file. If you have GaussView available to you, feel free to attempt this section. Otherwise, follow along to see what actions were taken. You have been provided a pdb file of the ligand that was present in the crystal structure in the product state. We will need to edit this structure to be in the reactant state.
The following steps were taken to prepare a Gaussian input file for O6 methylated guanine based on the crystal structure coordinates of the guanine product in the crystal. Recall that the methyl group had already been transferred from the ligand.
[user@local] gv GUN.pdb 
The input file would look something like this:
%chk=GUN.chk
# hf/3-21g
Title Card Required
0 1
N 20.05213100 11.86058800 36.02351400
C 20.78663800 12.65045600 36.89290400
N 20.90570600 12.13218500 38.09712300
C 20.20665200 10.92676800 38.02243900
C 19.93136600 9.86610100 38.92113800
O 20.33478900 9.77794100 40.20264300
N 19.21047500 8.81736400 38.51825700
C 18.75247500 8.79396500 37.24439200
N 17.96142200 7.71264700 36.91957400
N 18.94068400 9.71428900 36.27043500
C 19.66323200 10.73955800 36.72590700
H 19.82722600 12.05265800 35.05295000
H 21.20875900 13.60185700 36.57547400
H 17.88964300 7.52051800 35.92518000
H 18.08578800 6.90435000 37.52121100
C 21.11877500 10.85660800 40.73877700
H 20.56584100 11.80695700 40.70657800
H 21.32467200 10.57091700 41.77831600
H 22.06005100 10.98164000 40.18349700
In this section you will build and optimize the ligand structure and calculate corresponding electrostatic potential (ESP) charges using Gaussian in order to generating non-standard residue parameters. You will generate parameters for the non-standard O6 methylated guanine ligand using the restrained electrostatic potential (RESP) procedure. The input file was edited to run an optimization of the structure and an electrostatic potential (ESP) calculation. The optimization will be done at a high level of DFT, and the ESP will be computed at the Hartree Fock level to be consistent with the methods used to generate the standard AMBER charges.
%chk=GUN.chk
%nprocshared=4
%mem=10GB
# opt pbepbe/6-31g(2d,2p)
GUN_opt
0 1
N 20.05213100 11.86058800 36.02351400
C 20.78663800 12.65045600 36.89290400
N 20.90570600 12.13218500 38.09712300
C 20.20665200 10.92676800 38.02243900
C 19.93136600 9.86610100 38.92113800
O 20.33478900 9.77794100 40.20264300
N 19.21047500 8.81736400 38.51825700
C 18.75247500 8.79396500 37.24439200
N 17.96142200 7.71264700 36.91957400
N 18.94068400 9.71428900 36.27043500
C 19.66323200 10.73955800 36.72590700
H 19.82722600 12.05265800 35.05295000
H 21.20875900 13.60185700 36.57547400
H 17.88964300 7.52051800 35.92518000
H 18.08578800 6.90435000 37.52121100
C 21.11877500 10.85660800 40.73877700
H 20.56584100 11.80695700 40.70657800
H 21.32467200 10.57091700 41.77831600
H 22.06005100 10.98164000 40.18349700
--link1--
%chk=GUN.chk
%nprocshared=4
%mem=10GB
#P HF/6-31G* SCF(Conver=6) NoSymm Test Pop=mk IOp(6/33=2) guess=read geom=allcheck GFInput GFPrint
The first section indicates the optimization, and the link uses the optimized structure to calculate the ESP. The ESP will be represented on a grid of points. The keyword IOp(6/33=2) instructs Gaussian to output this information for use later when performing the RESP fit. You have also been provided a file called gau.slurm to run the job on expanse.
[user@cluster] sbatch gau.slurm When the job is finished, make sure the last line of GUN.log reports "Normal termination of Gaussian".
Next we will use the antechamber program to assign point charges based on the ESP. The fit will be performed such that topologically equivalent atoms have the same charge.
[user@cluster] antechamber -i GUN.log -fi gout -o GUN.mol2 -fo mol2 -c resp -at gaff -pf y -rn GUN This will create GUN.mol2. We used the flag -c resp to read the ESP from the Gaussian output file and perform RESP. The -at gaff flag means atoms types are assigned from the GAFF force field, -pf y removes intermediate files, and -rn GUN sets the residue name in the mol2 file to be GUN. This is the name we used in the PDB file when building the system.
@<TRIPOS>MOLECULE
GUN
19 20 1 0 0
SMALL
resp
@<TRIPOS>ATOM
1 N1 20.0520 11.8610 36.0240 na 1 GUN -0.334523
2 C1 20.7870 12.6500 36.8930 cc 1 GUN 0.097034
3 N2 20.9060 12.1320 38.0970 nd 1 GUN -0.523097
4 C2 20.2070 10.9270 38.0220 ca 1 GUN 0.144003
5 C3 19.9310 9.8660 38.9210 ca 1 GUN 0.558035
6 O1 20.3350 9.7780 40.2030 os 1 GUN -0.329698
7 N3 19.2100 8.8170 38.5180 nb 1 GUN -0.745822
8 C4 18.7520 8.7940 37.2440 ca 1 GUN 0.884237
9 N4 17.9610 7.7130 36.9200 nh 1 GUN -0.890772
10 N5 18.9410 9.7140 36.2700 nb 1 GUN -0.693379
11 C5 19.6630 10.7400 36.7260 ca 1 GUN 0.325075
12 H1 19.8270 12.0530 35.0530 hn 1 GUN 0.347262
13 H2 21.2090 13.6020 36.5750 h5 1 GUN 0.176097
14 H3 17.8900 7.5210 35.9250 hn 1 GUN 0.381855
15 H4 18.0860 6.9040 37.5210 hn 1 GUN 0.381855
16 C6 21.1190 10.8570 40.7390 c3 1 GUN -0.005320
17 H5 20.5660 11.8070 40.7070 h1 1 GUN 0.075719
18 H6 21.3250 10.5710 41.7780 h1 1 GUN 0.075719
19 H7 22.0600 10.9820 40.1830 h1 1 GUN 0.075719
@<TRIPOS>BOND
1 1 2 1
2 1 11 1
3 1 12 1
4 2 3 2
5 2 13 1
6 3 4 1
7 4 5 ar
8 4 11 ar
9 5 6 1
10 5 7 ar
11 6 16 1
12 7 8 ar
13 8 9 1
14 8 10 ar
15 9 14 1
16 9 15 1
17 10 11 ar
18 16 17 1
19 16 18 1
20 16 19 1
@<TRIPOS>SUBSTRUCTURE
1 GUN 1 TEMP 0 **** **** 0 ROOTThe mol2 file contains the point charges and atom types. For example, notice that the methyl group hydrogen atoms (H5,H6, and H7) were all assigned the same charge because they are topologically equivalent. When fitting was performed, it would have been done under the constraint that these should have the same charge. Similarly, the amine hydrogens (H3 and H4) are also equivalent.
[user@cluster] sed -n -e '9,27p' GUN.mol2 | awk '{print $9}' > charges.dat
[user@cluster] cat charges.dat
-0.334523
0.097034
-0.523097
0.144003
0.558035
-0.329698
-0.745822
0.884237
-0.890772
-0.693379
0.325075
0.347262
0.176097
0.381855
0.381855
-0.005320
0.075719
0.075719
0.075719
[user@cluster] awk '{ sum += $1 } END { print sum }' charges.dat
-1e-06Now we must create parameter files that are compatible with AMBER.
cat << EOF > tleap_GUN.in
source leaprc.gaff
GUN = loadmol2 GUN.mol2
SaveOff GUN GUN.lib
saveamberparm GUN GUN.parm7 GUN.rst7
quit
EOF
tleap -sf tleap_GUN.inWe will determine the geometric parameters from the GAFF force field. Each atom in the parameter file has been assigned a type from the GAFF force field, so we will search the existing library of parameters for these atom type.
cat << EOF > parmed_GUN.in
parm GUN.parm7
loadRestrt GUN.rst7
writeFrcmod GUN.frcmod
quit
EOF
parmed -i parmed_GUN.inNow you have the parameter files that were used to build the MTR1 system in the previous tutorial. The parameters for the protonated cytosine residue (CX) were generated in similar fashion.