This tutorial will demonstrate how to prepare a set of ligand-receptor complexes with coordinates in PDB format for simulation in AMBER using AMBERTools.
For this tutorial, we will prepare a set of ligands in complex with tyrosine kinase 2 (TYK2) for simulations in both the aqueous and ligand-receptor complex phases. The success of binding free energy calculations can be dependent on the initial pose of ligands in the receptor environment. As many ligands lack crystallographic data, there are two strategies commonly used to generate the initial structures [ Athanasiou 2017]. The first involves using a docking software, such as AutoDock Vina [ Eberhardt 2021], to generate an initial binding pose by docking the ligand into the receptor. The second involves mapping the ligands onto a chemically simular ligand for which there is an existing structure for the ligand-receptor complex. For this case, we will use the latter method. In this tutorial, we will prepare four ligands to be used in RBFE simulation a future tutorial. We will use ligands 31 and 42 from the Eur. J. Med. Chem article [ Liang 2013 1] hereon refered to as ejm_31 and ejm 42. We will aslo use lignads 27 and 28 from the J. Med. Chem. article [ Liang 2013 2], hereon refered to as jmc_27 and jmc_28. None of the four ligands (ejm_42, ejm_31, jmc_27, and jmc_28) have been crystalized in complex with TYK2; however, ligand 46 from the Eur. J. Med. Chem article is strucutally simular to the ligands to be used in this tutorial and does have crystallographic data (PDB: 4GIH).
To begin, let us prepare some directories. First, with-in the SmallMolecules directory prepared in the Acetic Acid tutorial, create a second directory called TYK2.
Inside the TYK2 directory, download the pdb structure for PDB: 4GIH using pdb4amber :
pdb4amber --pdbid 4GIH --dry --reduce -o 4gih.pdb
The --reduce option adds hydrogens to the protein, including residues that can have multiple protenation states. Please note that this may not be the best option for determining protenation state. More rigourous methods, such as using the H++ server may be advisable for applications outside of this tutorial.
We will rename the small molecule that we will use for atom mapping :
sed -i "s/0X5/LIG/g" 4gih.pdb
For our own convienence, we will also extract just the receptor to a pdb file:
sed -e "/LIG/d" 4gih.pdb > receptor.pdb
Both receptor.pdb and the final version of 4gih.pdb are available in the /expanse/projects/qstore/amber_ws/tutorials/pdb4amber/outputs directory
Next, we will will map our set of ligands onto the existing set of ligands. The sdf files for these ligands can be found here. the initial coordinate for each ligand come from the Open Force Field Inititive's protein-ligand-benchmark [ Hahn 2022]. For an example, we will preapre the coordinates for ejm_42. Create a directory called ligands but do not enter the directory. Open a python file, atom_map.py in a text editor and write the following :
from rdkit import Chemfrom rdkit.Chem import AllChemfrom rdkit.Chem import rdFMCSimport numpy as np# Load the PDB filepdb1 = Chem.MolFromPDBFile("4gih.pdb", removeHs=True)# Load the moleculesmol1 = Chem.SDMolSupplier("ejm_42.sdf")[0]# Extract the ligand from the PDBmol = Chem.MolFromPDBBlock("")for res in Chem.GetMolFrags(pdb1, asMols=True): if res.GetAtomWithIdx(0).GetPDBResidueInfo().GetResidueName() == "LIG": mol2 = res break# Find the Maximum Common Substructure (MCS)mcs = rdFMCS.FindMCS([mol1, mol2])common_smarts = mcs.smartsStringcommon_mol = Chem.MolFromSmarts(common_smarts)# Get the atom indices of the common scaffold in both moleculesmatch1 = mol1.GetSubstructMatch(common_mol)match2 = mol2.GetSubstructMatch(common_mol)# Align mol1 to mol2 based on the common scaffoldAllChem.AlignMol(mol1, mol2, atomMap=list(zip(match1, match2)))# Add Hydrogensmol1 = Chem.AddHs(mol1, addCoords=True)# Save the aligned molecule to a new PDB fileChem.MolToPDBFile(mol1, "ligands/ejm_42.pdb")The used python to run this script:
python3 atom_map.py
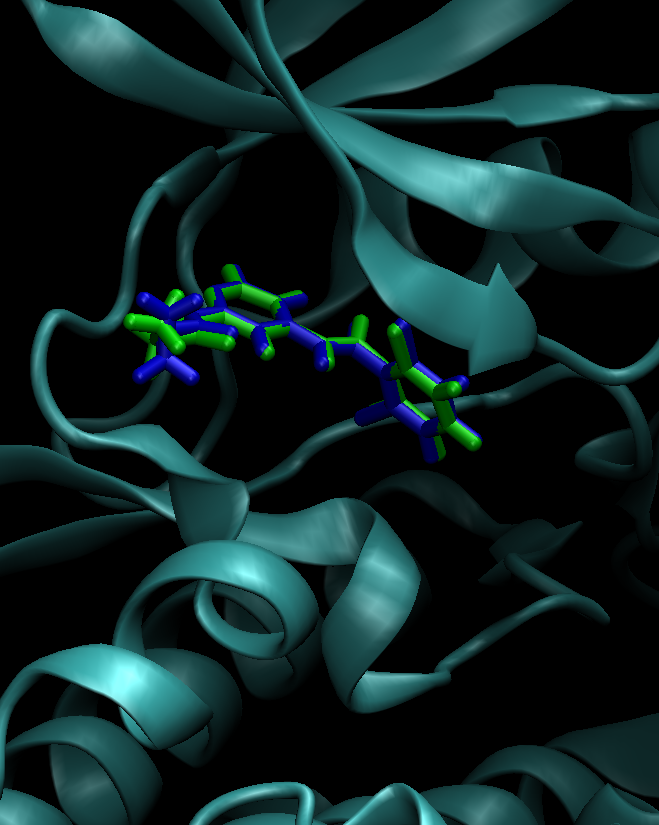
The aligned coordinates for ejm_42 will be written to ligands/ejm_42.pdb. This file is also available at /expanse/projects/qstore/amber_ws/tutorials/Param_tyk2/pdb4amber/outputs/ejm_42.pdb. Load both ligands/ejm_42.pdb and 4gih.pdb into VMD. As an exersize, verify that the two ligands (both using RESNAME LIG) are aligned (Fig. 1).

Before we can continue, we will need to find the atom charges and atom-types to be used in our simulations. To do this we will follow a procedure simular to that previously used in the acetic acid tutorial :
First, we will assign atomic charges and atom types and ouput the new charges, atom types, and coordinates to a mol2. The atom types will be assigned using the GAFF2 forcefield which is parameterized for RESP charges; consequently, we will assign chargers using the RESP method, which requires the Gaussian QM software package. We will do this in two steps. First, we will generate the inputfiles needed for Gaussian using antechamber:
antechamber -i ligands/ejm_42.pdb -fi pdb -o ejm_42.g16 -fo gcrt -nc 0 -gk 'p HF/6-31G(d) nosymm Pop=MK iop(6/33=2) iop(6/42=6) iop(6/50=1)' -gv 1 -ge ejm_42.resp -gn '%nproc=1' -gm '%mem=8000MB'
Now that we have the Gaussian files needed to calculate the RESP charges, load the Gaussian module, define the Gaussian scratch directory and pass the g16 file to Gaussian :
module purgemodule load cpu/0.15.4module load gaussian/16.C.01GAUSS_SCRDIR=/scratch/$USER/job_$SLURM_JOBIDg16 ejm_42.g16 > ejm_42.log
If Gaussian is unavailable, the log file is available at /expanse/projects/qstore/amber_ws/tutorials/Param_tyk2/antechamber/intermediates_io.
Once Gaussian has completed the calculation, reload the amber modules and use antechamber to extract the charges from the log file and assign atom types:
module load workshop/software
antechamber -i ejm_42.log -fi gout -gv 0 -o ejm_42.mol2 -fo mol2 -c resp -nc 0 -rn LIG -at gaff2
The mol2 file is available in the /expanse/projects/qstore/amber_ws/tutorials/Param_tyk2/antechamber/outputs.
Now that we have the mol2 file, we can generate the frcmod file using the parmchk2 executable:
parmchk2 -i ejm_42.mol2 -o ejm_42.frcmod -f mol2 -s 2
Like antechamber, you can learn more about the executable options for parmchk2 using "parmchk2 -h". The ejm_42.frcmod file is available at /expanse/projects/qstore/amber_ws/tutorials/Param_tyk2/parmchk2/outputs
Next, we will use the mol2 and frcmod files to generate our AMBER formated parameters and restart files. We will solvate the sytem using the OPC water model. Open a file ejm_42_aq_tleap.in in a text editor and write the following :
source leaprc.protein.ff19SB
loadamberparams frcmod.ff19SB
source leaprc.water.opc
loadamberparams frcmod.opc
source leaprc.gaff2
loadamberparams ejm_42.frcmod
LIG = loadmol2 "ejm_42.mol2"
saveOff LIG ejm_42.lib
solvateOct ejm_42 OPCBOX 20.0
saveAmberParm ejm_42 ejm_42_aq.parm7 ejm_42_aq.rst7
quit
Then run the command:
tleap -f ejm_42_aq_tleap.in
Next, we will prepare the protein ligand complex systems :
source leaprc.protein.ff19SB
loadamberparams frcmod.ff19SB
source leaprc.water.opc
loadamberparams frcmod.opc
source leaprc.gaff2
loadamberparams ejm_42.frcmod
ejm_42_lig = loadmol2 "ejm_42.mol2"
ejm_42_rec = loadpdb "receptor.pdb"
complex = combine {ejm_42_lig ejm_42_rec}
savepdb complex ejm_42.pdb
addIons complex Na+ 0
solvateOct complex OPCBOX 16.0
saveAmberParm complex ejm_42_com.parm7 ejm_42_com.rst7
quit
Then run the command:
tleap -f ejm_42_com_tleap.in
Because the protein carries a net charge of -2, sodium counter ions are added. The parm7, rst7 and lib files are available at /expanse/projects/qstore/amber_ws/tutorials/Param_tyk2/tleap/outputs
Repeat this process for the remaining ligands. This can be automated by running the shell script available at /expanse/projects/qstore/amber_ws/tutorials/Param_tyk2/misc/run.sh