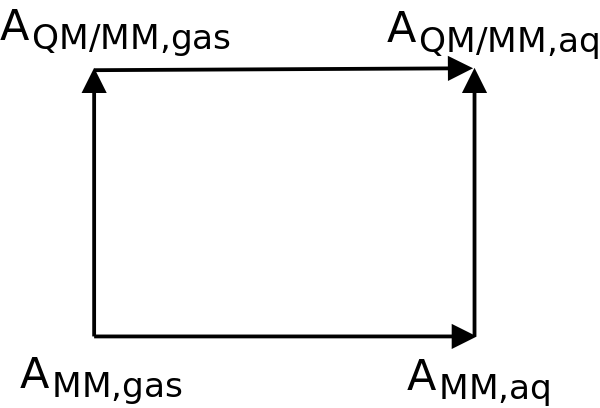
This tutorial will demonstrate how to calculate the absolute solvation free energy of a small molecule using a QM/MM potential energy function. Specifically, we shall estimate the solvation free energy of acetic acid using the GFN2-xTB semiempirical model supplemented with a QDπ2 machine learning potential. The method of calculation is based on the use of an indirect thermodynamic cycle, illustrated in Figure 1.

It is impractical to directly estimate the solvation free energy using a QM/MM potential [line 1 of Eq. \eqref{E:dAQMMM}] due to its high cost and the large amount of required sampling. \begin{equation}\label{E:dAQMMM} \begin{split} \Delta A_{\text{QM/MM},\text{solv}} =& A_{\text{QM/MM},\text{aq}} - A_{\text{QM/MM},\text{gas}} \\ =& \Delta A_{\text{MM},\text{solv}} + \Delta A_{\text{MM}\rightarrow\text{QM/MM},\text{aq}} - \Delta A_{\text{MM}\rightarrow\text{QM/MM},\text{gas}} \end{split} \end{equation} In contrast, a MM force field is sufficiently affordable to obtain reasonable estimates of ΔAMM,solv [Eq. \eqref{E:dAMM}]. \begin{equation}\label{E:dAMM} \Delta A_{\text{MM},\text{solv}} = A_{\text{MM},\text{aq}} - A_{\text{MM},\text{gas}} \end{equation} Because the free energy is a state function, the sum of free energies along any closed path is zero. This is mathematically expressed by the second line in Eq. \eqref{E:dAQMMM}. In other words, the indirect thermodynamic approach calculates ΔAQM/MM,solv from a MM reference potential by including "bookend corrections" [Eqs. \eqref{E:BEaq}-\eqref{E:BEgas}]. \begin{equation}\label{E:BEaq} \Delta A_{\text{MM}\rightarrow\text{QM/MM},\text{aq}} = A_{\text{QM/MM},\text{aq}} - A_{\text{MM},\text{aq}} \end{equation} \begin{equation}\label{E:BEgas} \Delta A_{\text{MM}\rightarrow\text{QM/MM},\text{gas}} = A_{\text{QM/MM},\text{gas}} - A_{\text{MM},\text{gas}} \end{equation} The bookend corrections often do not require extensive sampling because it does not involve a physical transformation of the system.
A bookend correction can be calculated from the thermodynamic integration (TI) [Eq. \eqref{E:TI}] or multistate Bennett acceptance ratio method (MBAR) by analyzing the equilibrium sampling produced by a λ-dependent potential energy function that transforms between MM and QM/MM potentials [Eq. \eqref{E:Ulam}]. \begin{equation}\label{E:TI} \Delta A_{\text{MM}\rightarrow\text{QM/MM}} = \int_{0}^{1} \left\langle \frac{\partial U}{\partial \lambda} \right\rangle_{\lambda} d\lambda \end{equation} \begin{equation}\label{E:Ulam} U(\mathbf{r};\lambda) = \lambda U_{\text{QM/MM}}(\mathbf{r}) + (1-\lambda) U_{\text{MM}}(\mathbf{r}) \end{equation} \begin{equation}\label{E:dUdlam} \frac{\partial U(\mathbf{r};\lambda)}{\partial\lambda} = U_{\text{QM/MM}}(\mathbf{r}) - U_{\text{MM}}(\mathbf{r}) \end{equation}
"Multisander" (i.e., the MPI parallel version of sander, sander.MPI) evaluates Eq. \eqref{E:Ulam} using a "groupfile" and a few key options in the &cntrl; namelist. The first line of the groupfile defines the MM potential energy (the λ=0 state), and the second defines the QM/MM potential energy (the λ=1 state). An example is shown in Figure 2.
-O -p ../../../common/aq.parm7 -i mm.0.00.mdin -o mm.0.00.mdout -c init.0.00.rst7 -r mm.0.00.rst7 -x mm.0.00.nc -inf mm.0.00.mdinfo
-O -p ../../../common/aq.parm7 -i qm.0.00.mdin -o qm.0.00.mdout -c init.0.00.rst7 -r qm.0.00.rst7 -x qm.0.00.nc -inf qm.0.00.mdinfo
The MM and QM/MM input files must be the exact same except for the activation/deactivation of the QM (and machine learning) potentials. The mdin file &cntrl; section must specify the following options:
icfe=1
This causes the groupfile to be interpretted as
defining a λ-dependent potential energy function.
clambda=0.0
This is the value of λ (a floating point number between 0 and 1).
The λ=0 state corresonds to the first line in the groupfile.
Both the MM and QM/MM input files should specify the same clambda value.
The following is a list of options that may be different in the MM and QM/MM input files.
ifqnt=0
The value of this option (appearing in the &cntrl; section)
controls whether a QM/MM potential is applied.
idprc=0
The value of this option (appearing in the optional &dprc; section)
controls whether a DeePMD-kit machine learning correction is applied.
The &dprc; section is only applicable if amber was configured
with interface support to the DeePMD-kit library.
qm_ewald=0
The value of this option (appearing in the &qmmm; section)
controls whether periodic electrostatics are included in the
QM calculation. One normally sets qm_ewald=0; however, when
calculating a bookend calculation in the gas phase, one can
turn off the periodic electrostatics.
Example MM and QM/MM inputs for the calculation of Eq. \eqref{E:BEaq} are shown in Figure 3.
Title: MM input files
&cntrl
! IO =======================================
irest = 1 ! 0 = start, 1 = restart
ntx = 5 ! 1 = start, 5 = restart
ntxo = 1 ! rst is formatted file
iwrap = 1 ! wrap crds to unit cell
ioutfm = 1 ! write mdcrd as netcdf
ntpr = 10 ! mdout print freq
ntwx = 0 ! mdcrd print freq
ntwr = 500 ! rst print freq
ntwv = 0
! DYNAMICS =================================
imin = 0 ! Run dynamics
dt = 0.002! ps/step
nstlim = 2500 ! number of time steps
ntb = 1 ! 1=periodic box
! TEMPERATURE ==============================
temp0 = 298 ! target temp
gamma_ln = 5.0 ! Langevin collision freq
ntt = 3 ! thermostat (3=Langevin)
! PRESSURE ================================
ntp = 0 !0=off 1=isotropic scaling
! SHAKE ====================================
ntc = 2
ntf = 1
noshakemask=':1'
! MISC =====================================
cut = 10
ig = -1
ifqnt = 0
! TI =======================================
icfe = 1 ! FE simulation
clambda = 0.0 ! lambda. 0=MM, 1=QM/MM
ifmbar = 1 ! print mbar energies
bar_intervall= 10 ! print freq
bar_l_min = 0
bar_l_max = 1
bar_l_incr = 1
dynlmb = 0
ntave = 0
/
&qmmm
qm_theory = 'XTB'
qmmask = ':1'
qmcharge = 0
spin = 1
qmshake = 0
qm_ewald = 1
qmmm_switch = 1
scfconv = 1.e-10
verbosity = 0
tight_p_conv = 1
diag_routine = 0
pseudo_diag = 1
dftb_maxiter = 100
/
&xtb
qm_level="GFN2-xTB"
/
&dprc
idprc=0
mask=":1"
rcut=0.0
intrafile(1) = "../../../common/qdpi2.pb"
/
Title: QM/MM+MLP input file
&cntrl
! IO =======================================
irest = 1 ! 0 = start, 1 = restart
ntx = 5 ! 1 = start, 5 = restart
ntxo = 1 ! rst is formatted file
iwrap = 1 ! wrap crds to unit cell
ioutfm = 1 ! write mdcrd as netcdf
ntpr = 10 ! mdout print freq
ntwx = 0 ! mdcrd print freq
ntwr = 500 ! rst print freq
ntwv = 0
! DYNAMICS =================================
imin = 0 ! Run dynamics
dt = 0.002! ps/step
nstlim = 2500 ! number of time steps
ntb = 1 ! 1=periodic box
! TEMPERATURE ==============================
temp0 = 298 ! target temp
gamma_ln = 5.0 ! Langevin collision freq
ntt = 3 ! thermostat (3=Langevin)
! PRESSURE ================================
ntp = 0 !0=off 1=isotropic scaling
! SHAKE ====================================
ntc = 2
ntf = 1
noshakemask=':1'
! MISC =====================================
cut = 10
ig = -1
ifqnt = 1
! TI =======================================
icfe = 1 ! FE simulation
clambda = 0.0 ! lambda. 0=MM, 1=QM/MM
ifmbar = 1 ! print mbar energies
bar_intervall= 10 ! print freq
bar_l_min = 0
bar_l_max = 1
bar_l_incr = 1
dynlmb = 0
ntave = 0
/
&qmmm
qm_theory = 'XTB'
qmmask = ':1'
qmcharge = 0
spin = 1
qmshake = 0
qm_ewald = 1
qmmm_switch = 1
scfconv = 1.e-10
verbosity = 0
tight_p_conv = 1
diag_routine = 0
pseudo_diag = 1
dftb_maxiter = 100
/
&xtb
qm_level="GFN2-xTB"
/
&dprc
idprc=1
mask=":1"
rcut=0.0
intrafile(1) = "../../../common/qdpi2.pb"
/
idprc=1
turns on the machine learning correction, whose
parameters are read from the file listed in
intrafile(1).
The machine learning potential is applied to the
atoms in mask, which is the selected
QM atoms. The QDπ2 model corrects the intra-molecular
interactions (not the QM/MM interactions); therefore,
rcut=0.0.
Because the potential energy is described using a groupfile,
one needs the MPI version of sander and at least 2 CPU cores.
On a slurm-based scheduling system, one can launch 2 (MPI) tasks
on 1 node using 1 core per task with the following command, which
one would write in their submission slurm script.
The -ng 2 tells sander to expect to read 2 lines
from the groupfile.
srun -N 1 -n 2 -c 1 --mpi=pmi2 sander.MPI -ng 2 -groupfile mm2qmmm.0.00.groupfile
For accurate results, one should simulate several λ values, such as 0.0, 0.2, 0.4, 0.6, 0.8, and 1.0. The bookend transformations tend to require sampling of far fewer λ-states than alchemical transformations because the system is not undergoing a physical change. The free energy calculated from TI or MBAR requires equilibrium statistics; therefore, one should equilibrate each simulation before gathering production statistics.
tar -xzf example_mm2qmmm.inputs.tar.gz.
This will create a directory ./example_mm2qmmm/.
./example_mm2qmmm/
and type bash RunExample.sh.
The RunExample.sh script will prepare a series of
groupfiles and corresponding MM and QM/MM input files.
These files are organized into several subdirectories, named:
prod01, prod02, prod03, prod04.
These are 4 directories are used to perform independent trials
that depart from structures extracted from
MM simulations.
The trial directories will contain 2 subdirectories:
aq/ and vac which contain simulations
for Eqs. \eqref{E:BEaq} and \eqref{E:BEgas}, respectively.
The sampling within each environment is separated into two directories:
equil/ and prod/,
which are used to perform equilibration and production sampling,
respectively.
prod01/prod02/prod03/prod04/template/aq/
vac/
equil/
prod/
mm2qmmm.*.groupfile
mm.*.mdin
qm.*.mdin
RunExample.sh script.
The highlighted files and directories are created by the script.
The prod01, prod02, prod03, prod04 are 4 independent trials
starting from different snapshots extracted from
previous MM simulations. These directories are copies of the
template directory.
RunExample.sh script will prepare a sub.sims.slurm
script. The submission script uses slurm arrays to perform the
sampling in each trial directory. In other words, the 4 array values
(#SBATCH --array=1-4) perform sampling in each
of the 4 trial directories.
prod*/*/prod/mm.*.mdout.
For your convenience, you can download the necessary output files
example_mm2qmmm.outputs.tar.gz.
Unpacking the files with tar -xzf example_mm2qmmm.outputs.tar.gz
will cause the files to be copied into the subfolder ./example_mm2qmmm.
That is, unpack example_mm2qmmm.outputs.tar.gz in the same directory you unpacked
example_mm2qmmm.inputs.tar.gz
analysis/prod01/prod02/prod03/prod04/
aq/
vac/
equil/
prod/
mm.*.mdout
The MM and QM/MM mdout files contain the same potential energy and the ∂U/∂λ data; therefore, we will only look at the MM mdout files. To perform TI analysis, one must extract the ∂U/∂λ time series from the simulations of each λ-state in each environment (aq or vac) and trial (prod01, prod02, prod03, prod04). Similarly, MBAR analysis requires the potential energy of each state, U(r;λ'). Because this is a linear transformation, the potential energy of any intermediate state can be computed from the λ=0 and λ=1 states [Eq. \eqref{E:Ulam}]. Furthermore, the MBAR analysis only requires potential energy differences, so we will obtain the same free energy if -- for each frame, k -- we define the zero of energy to be UMM(rk). In which case, the potential energy of state λ' evaluated at frame k within the ensemble of state λ is given by Eq. \eqref{E:UMBAR}. \begin{equation}\label{E:UMBAR} \begin{split} U(\mathbf{r}_{\lambda k};\lambda^{\prime}) - U_{\text{MM}}(\mathbf{r}_{\lambda k}) = & \lambda^{\prime} \left[ U_{\text{QM/MM}}(\mathbf{r}_{\lambda k})-U_{\text{MM}}(\mathbf{r}_{\lambda k}) \right] \\ =& \lambda^{\prime} \frac{\partial U(\mathbf{r}_{\lambda k};\lambda)}{\partial \lambda} \end{split} \end{equation} The mdout files contain lines that look like:
NSTEP = 50 TIME(PS) = 1020.100 TEMP(K) = 298.61 PRESS = 0.0
Etot = -61089.2946 EKtot = 5235.5357 EPtot = -66324.8304
BOND = 2.8694 ANGLE = 4.6157 DIHED = 17.6568
1-4 NB = 0.4164 1-4 EEL = -89.3041 VDWAALS = 5313.5873
EELEC = -42803.8431 EHBOND = 0.0000 RESTRAINT = 0.0000
DV/DL = -143769.6429
Ewald error estimate: 0.2471E-02
------------------------------------------------------------------------------example_mm2qmmm/prod01/aq/prod/mm.0.20.mdout.
That is, it is sample from the λ=0.2 ensemble within the first trial performed in solution.
The ∂U/∂λ value is -143769.6429 kcal/mol (the number after DV/DL).
The example_mm2qmmm/sub.ana.slurm slurm submission file uses
the
example_mm2qmmm/edgembar-bookend2dats.py
script to extract the relevant data from the mdout files and use edgembar
to perform the analysis.
The "bookend-to-dats" script takes a list of mdout filenames
(e.g. prod01/aq/prod/mm.*.mdout)
extracts the DV/DL data from each file,
and writes the TI and MBAR timeseries in a format
that can be analyzed using the edgembar program.
The MakeAnalysis.sh script
executes the script on the mdout files
from each trial.
The extracted outputs are written to a series of files:
analysis/mm~qmmm/env/trial/dats/efep_tlam_elam.dat
and
analysis/mm~qmmm/env/trial/dats/dvdl_tlam.dat,
where env is either aq or vac,
trial is either t01, t02, t03, or t04,
tlam is the lambda value used to simulate the trajectory,
and elam is the lambda value corresponding to the energy within the file.
The sub.ana.slurm script
then creates an XML input file to run edgembar.
The XML file is provided as mm2qmmm/analysis/xml/mm~qmmm.xml,
and it is shown in Figure 7.
<?xml version="1.0" ?>
<edge name="mm~qmmm">
<env name="target">
<stage name="STAGE">
<trial name="t01">
<dir>mm~qmmm/aq/t01/dats</dir>
<ene>0.00000000</ene>
<ene>0.20000000</ene>
<ene>0.40000000</ene>
<ene>0.60000000</ene>
<ene>0.80000000</ene>
<ene>1.00000000</ene>
</trial>
<trial name="t02">
<dir>mm~qmmm/aq/t02/dats</dir>
<ene>0.00000000</ene>
<ene>0.20000000</ene>
<ene>0.40000000</ene>
<ene>0.60000000</ene>
<ene>0.80000000</ene>
<ene>1.00000000</ene>
</trial>
<trial name="t03">
<dir>mm~qmmm/aq/t03/dats</dir>
<ene>0.00000000</ene>
<ene>0.20000000</ene>
<ene>0.40000000</ene>
<ene>0.60000000</ene>
<ene>0.80000000</ene>
<ene>1.00000000</ene>
</trial>
<trial name="t04">
<dir>mm~qmmm/aq/t04/dats</dir>
<ene>0.00000000</ene>
<ene>0.20000000</ene>
<ene>0.40000000</ene>
<ene>0.60000000</ene>
<ene>0.80000000</ene>
<ene>1.00000000</ene>
</trial>
</stage>
</env>
<env name="reference">
<stage name="STAGE">
<trial name="t01">
<dir>mm~qmmm/vac/t01/dats</dir>
<ene>0.00000000</ene>
<ene>0.20000000</ene>
<ene>0.40000000</ene>
<ene>0.60000000</ene>
<ene>0.80000000</ene>
<ene>1.00000000</ene>
</trial>
<trial name="t02">
<dir>mm~qmmm/vac/t02/dats</dir>
<ene>0.00000000</ene>
<ene>0.20000000</ene>
<ene>0.40000000</ene>
<ene>0.60000000</ene>
<ene>0.80000000</ene>
<ene>1.00000000</ene>
</trial>
<trial name="t03">
<dir>mm~qmmm/vac/t03/dats</dir>
<ene>0.00000000</ene>
<ene>0.20000000</ene>
<ene>0.40000000</ene>
<ene>0.60000000</ene>
<ene>0.80000000</ene>
<ene>1.00000000</ene>
</trial>
<trial name="t04">
<dir>mm~qmmm/vac/t04/dats</dir>
<ene>0.00000000</ene>
<ene>0.20000000</ene>
<ene>0.40000000</ene>
<ene>0.60000000</ene>
<ene>0.80000000</ene>
<ene>1.00000000</ene>
</trial>
</stage>
</env>
</edge>analysis/ directory.
The name of the edge is "mm~qmmm".
There are two environments: "target" is the solvated phase and "reference"
is the gas phase.
The free energy simulations involved a single stage, which we simply name "STAGE".
edgembar --no-auto ./xml/mm~qmmm.xml. The output will be written
to ./xml/mm~qmmm.py. (The output is a python script.)
One can view the output as an HTML file by running python3 ./xml/mm~qmmm.py,
which will produce ./xml/mm~qmmm.html.
One can view the free energy report here.
The reported free energy difference, ΔΔG
-- technically, the
calculated free energies are Helmholtz free energies because we performed
the bookend simulations at constant volume, but edgembar doesn't know that --
is the bookend free energy that one would use to correct the MM-calculated solvation
free energy.
A
guide for understanding the free energy report
is provided by the
edgembar
online documentation.